
This command opens a property sheet for defining the mechanism and the values of thermodynamic/kinetic parameters used by the fitting procedure as starting values. When closing the dialog with OK the Run command is automatically executed and the current curve is simulated on the basis of the entered mechanism and parameters using the experimental parameters imported from the active experiment. The simulated current curve is then shown in the client area together with the experimental counterpart. This enables the user to estimate the quality of mechanism and starting parameters.
Selecting Parameters for being optimized
Before the Data Fitting can be started it is necessary to select the parameters which are to be optimized. In the case of the CV-example project (contained in the file FittingExample.dep installed in DigiElch's home directory) the mechanism and starting parameters are defined in the following way
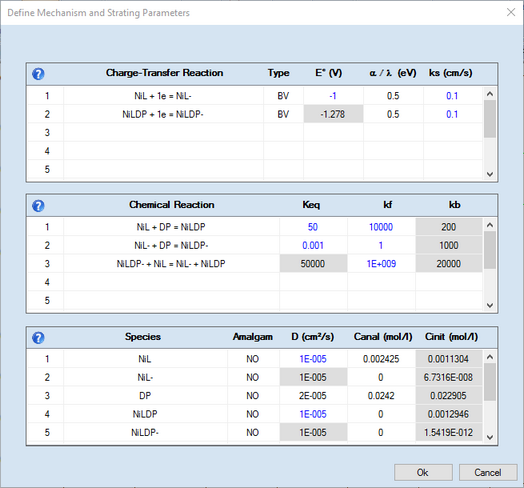
The refer to CV-experiments measured for a nickel(II)-chelate complex (denoted as NiL) in the absence and presence of dipyridine (denoted as DP).
 Note that the parameters selected to be optimized by the fitting procedure are plotted in blue. The value of an heterogeneous rate constant, ks, will be plotted in magenta if it is not only selected for being optimized but also linked to the surface coverage of a species.
Note that the parameters selected to be optimized by the fitting procedure are plotted in blue. The value of an heterogeneous rate constant, ks, will be plotted in magenta if it is not only selected for being optimized but also linked to the surface coverage of a species.
Fitting Charge Transfer Parameters
Click the right mouse button on the (charge-transfer reaction containing a) parameter value which is to be optimized. This opens the following dialog box
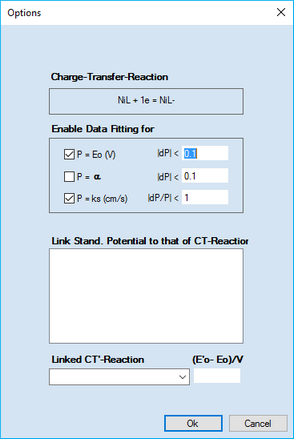
It can be used for selecting or de-selecting the three parameters associated with the underlying charge-transfer reaction. In the example on the left hand site, the standard potential, E°(V), and the heterogeneous rate constant, ks (cm/s), associated with the Charge Transfer Reaction NiL + e = NiL- will be optimized. Moreover, the alteration of the standard potential is restricted to 0.1 V per iteration and the relative change of ks remains smaller than the actual parameter value itself in each iteration.
Moreover, the standard potentials of other CT-Reactions (denoted by a prime) can be linked to that being optimized. In the above example the list of such CT'-Reactions (headlined Link Stand. Potential to that of CT-Reaction) is empty because only CT-Reactions not being fitted themselves and not determined by the thermodynamic constants of other reactions (thermodynamically superfluous reactions, TSR, indicated by a "blocked" grayed input field ) will be listed here. If this list is not empty any CT-Reaction selected there will be copied to the combo-box headlined Linked CT'-Reaction. The latter is used then for entering (individually for each reaction) the difference between the standard potential, E'0, referring to the linked CT-Reaction and E0 referring to NiL + e = NiL. The difference will not be altered in the course of Data Fitting.
Fitting Homogeneous Chemical Reaction Parameters
Click the right mouse button on the (homogeneous chemical reaction containing a) parameter value which is to be optimized. This opens the following dialog box
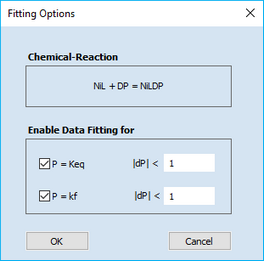
It can be used for selecting or de-selecting the two parameters associated with the underlying chemical reaction. In the above example, the equilibrium constant, Keq, and the (forward) rate constant, kf, associated with the Chemical Reaction, NiL + DP = NiLDP, will be optimized. Moreover, the relative alteration of both parameters remains smaller than the actual parameter values themselves in each iteration.
Fitting the diffusion coefficient of a species
Click the right mouse button on the value of the diffusion coefficient for a particular species. This opens the following dialog box
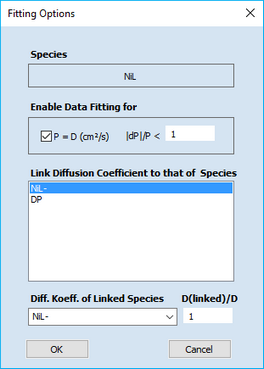
It can be used for selecting or deselecting the diffusion-coefficient associated with the underlying species.
The value of |dP|/P defines the maximum relative change of the parameter (diffusion coefficient) value permitted per iteration.
In addition, the dialog box provides the option for linking the diffusion coefficients of two or more species. This feature is very important. Because of the well-known relationship
E1/2=E° + (RT/nF) ln( DR/DO )
is it impossible to distinguish a shift of the standard potential from the potential shift originating from changing the ratio DR/DO . This leads to a perfect parameter coupling that cannot be unraveled mathematically. Consequently, the fitting procedure may not converge if a user is trying to fit the standard potential of a redox couple together with the diffusion coefficients of both species. Fitting the standard potentials makes sense only if the diffusion coefficients of all species involved in charge transfer reactions are explicitly known or if the ratio DR/DO for each redox couple is forced to remain constant during the data fitting. In the above dialog box the linking for DNiL and DNiLensures that DNiL/DNiL- = 1. Looking at the property sheet on the top of this page reveals that DNiLDP and DNiLDP- are also linked while DNiLDP and DNiL are to be optimized (plotted in blue). Due to the parameter linking the values of DNiL- and DNiLDP- cannot be modified independently of DNiL and DNiLDP . The input fields associated with linked parameters are therefore blocked as indicated by the gray background color.
